Submitted by Vicky Yee K Reid on Wed, 11/12/2024 - 10:38
The sustainable transformation of the chemical and pharmaceutical industry requires greener manufacturing. The use of enzymes as catalysts offers a promising route to less waste generation and lower energy consumption. However, such biocatalysts often require challenging engineering, where the combination of droplet microfluidics, deep sequencing and AI can make a difference.
A collaboration between the Hollfelder Lab (Biochemistry) and Lio Lab (Computer Science) at Cambridge, and the Mutti Lab at the University of Amsterdam, our team — comprising Maximilian Gantz, Simon Mathis, myself (Friederike Nintzel), and many others — has developed a new platform that integrates droplet microfluidics, deep sequencing and AI to accelerate the engineering of biocatalysts for greener production processes in the chemical and pharmaceutical industry.
Greener chemistry with biocatalysis
The chemical industry is the largest fossil fuel consumer, while the pharmaceutical industry emits more greenhouse gases than its automotive counterpart. To combat the climate crisis, these sectors must transition to more sustainable practices. One route is by improving process efficiencies and decreasing waste streams. A notable example is the US-American pharma firm MSD (Merck & Co) which recently innovated the production process of their HIV drug Islatravir by implementing a five-enzyme biocatalytic cascade, thereby cutting waste by 14-fold. Biocatalysis uses the natural diversity of enzymes and employs it for synthetic chemistry. Enzymes inherently offer stereoselectivity and are compatible with cascade and one-pot reactions. Therefore, biocatalytic reactions benefit from less waste generation and lower energy demand.
Efficient enzyme engineering is the key to putting biocatalysts into practice
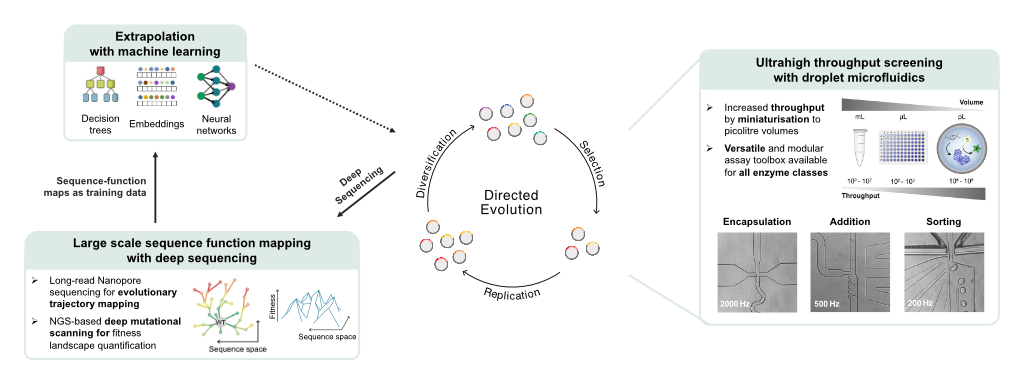
Natural enzymes are rarely efficient and stable enough for industrial application. Thus, protein engineers, like us in the Hollfelder lab, are needed to bridge this gap. Following the process of directed evolution, we generate large libraries of enzyme variants in the lab and test each enzyme to identify the most improving mutations. These are selected, replicated, and the cycle is repeated until the biocatalyst meets the desired characteristics. Unfortunately, beneficial mutations are rare and conventional directed evolution remains a labour-intensive hit-and-miss approach.
Combining droplet microfluidics, deep sequencing and AI to accelerate enzyme engineering
In the Hollfelder lab, we are tackling the challenge of identifying improving mutations. We use droplet microfluidics – scaling down enzyme reactions to pico-litre scale – to test more than a million reactions in as little as an hour. We match this throughput in experimentation with the throughput of NGS and Nanopore Sequencing. Thereby, we can not only test one million enzyme variants in an hour but also record their sequence identity and their activity. This allows us to build huge sequence-function datasets that reveal which enzyme regions are most receptive to mutations and how multiple mutations interact, even across long ranges. In collaboration with the Lio lab, we went one step further and used these datasets to train machine learning models and predict even better mutations. We could show that our experimental data is essential to teach models which know the general protein language about the idiosyncrasies that each enzyme carries in a particular reaction context. This demonstrates that our novel platform generates unique protein-specific data – not available from sources like PDB or AlphaFold – which is crucial for accurate AI predictions. Finally, and in collaboration with the Mutti lab, we applied our new technology to engineer a biocatalyst needed for sustainable production processes in the pharmaceutical industry.
Figure one: Data-driven directed evolution of biocatalysts at ultrahigh throughput
Further Information
For further information, please see our recent preprint and Faraday Discussions Review.
Credits & Acknowledgements
Credits to the whole team behind this research: Maximilian Gantz, Simon V. Mathis, Friederike E. H. Nintzel, Paul J. Zurek, Tanja Knaus, Elie Patel, Daniel Boros, Friedrich-Maximilian Weberling, Matthew R. A. Kenneth, Oskar J. Klein, Elliot J. Medcalf, Jacob Moss, Michael Herger, Tomasz S. Kaminski, Francesco G. Mutti, Pietro Lio, Florian Hollfelder and many more!