
Submitted by Jenny Molloy on Tue, 29/11/2016 - 12:59
In vitro or cell-free synthetic biology is a topic of growing interest to many groups in Cambridge and the Synthetic Biology SRI is pleased to announce an upcoming programme of activities to promote and support interdisciplinary work in this space.
In vitro synthetic biology allows programming of cell-free extracts from bacteria or other organisms with DNA-encoded ‘logic gates’ that respond in different ways to particular combinations of inputs or provide quantitative results.
Recent work combining this emerging technology with paper-based microfluidics has delivered rapid, low-cost paper tests for Ebola and Zika Virus and small molecule sensors such as glucose assays [1], which are stable in dried form for at least one year. There is no genetic modification involved and a fully equipped lab is not required once the cell-free extract is produced and stably dried down, making this technology far more accessible to researchers in low-resource settings than in vivo synthetic biology and vastly lowering the initial cost of engineering biosensors.
Kick-Off Event - SynBio Forum
The Synthetic Biology SRI has a range of activities over 2016-17 promoting cell-free systems as tools for research and education. This kicked off on 19 Oct 2016 with a SynBio Forum and workshop providing an overview of in vitro systems and their potential applications delivered by invited speaker Vincent Noireaux (University of Minnesota), who described his lab's work providing a toolbox for working with E.coli cell extracts [2].
 Nick Rollings of Cambridge Consultants then introduced XylemDx, their inkjet-printed, paper-based microfluidics and electronics solution for diagnostic sensors. Low-cost, disposable and easily fabricated sensors are a major technological opportunity that increase the potential for field applications of cell-free systems.
Nick Rollings of Cambridge Consultants then introduced XylemDx, their inkjet-printed, paper-based microfluidics and electronics solution for diagnostic sensors. Low-cost, disposable and easily fabricated sensors are a major technological opportunity that increase the potential for field applications of cell-free systems.
Fernan Fedirici (Pontificia Universidad Católica de Chile) introduced work in progress in his lab to use cell-free systems for exploring morphological patterning as well as lowering the cost of the associated hardware and biological reagents.
 A hands-on prototyping workshop was held the same day in the Plant Sciences Teaching Lab, led by Vincent Noireaux and Fernan Fedirici. Just over 20 early career researchers from across all five schools took part, several finding themselves in the lab for the first time.
A hands-on prototyping workshop was held the same day in the Plant Sciences Teaching Lab, led by Vincent Noireaux and Fernan Fedirici. Just over 20 early career researchers from across all five schools took part, several finding themselves in the lab for the first time.
Demonstrating the power of the system for teaching, the entire practical took just a few hours and delivered exactly the quantitative results predicted for all teams. The opportunity to programme complex logic circuits in reasonably short periods of time allows for a radical rethink of practical opportunities, including incorporation of the full design-build-test cycle that is so central to synthetic biology approaches.
Development i-Teams highlight impactful applications

Several of the workshop attendees, including an economist and philosopher, were members of a Development i-Team who looked for six weeks at applications of cell-free synthetic biology in diagnostics to improve the welfare of the bottom billion. The Development i-Teams programme is run by the Centre for Global Equality and the Institute for Manufacturing and the team were supported by Fernan Fedirici and Richard Hammond (Cambridge Consultants) to survey uses of the technology in several domains.
They presented their findings to an impressive audience on 22 November and selected the following applications for further exploration:
- Water contamination testing - heavy metals, often caused by mining operations.
- Crop disease - Cassava brown streak disease, a virus that threatens this key African food security crop and causes root damage that is not visible until harvest with an estimated $1Bn/year damage.
- Animal disease - Newcastle disease. A viral disease of farmed and wild poultry with estimated $2 billion annual impact.
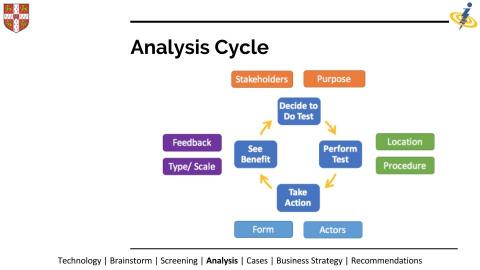
A key output of the project was an analysis cycle for assessing the suitability of different applications. This not only included technical feasibility but also actionable steps that could be taken post-diagnosis. This could include the availability of medication or remediation methods but also the context of the situation, such as local politics, power imbalances and more. The team then took a fascinating look at each of their chosen applications in the context of this analysis. Several members of the team now plan to continue exploring and developing applications, with the support of the Centre for Global Equality and the Synthetic Biology SRI.
Follow-on Activities
Interdisciplinary teams interested in cell-free systems are encouraged to apply to the OpenPlant Fund by 1 Dec 2016. This special theme is an addition to the Fund, which typically offers £5000 grant for innovative, open and interdisciplinary projects in plant synthetic biology. Examples of challenges that could be tackled include but are not limited to:
- Low-cost instrumentation for quantifying marker expression.
- Improving substrates for ‘paper-based’ assays.
- Developing lower-cost, more accessible cell extract production methods.
- Methods for normalising results across multiple experiments and batches of extract.
- Prototyping interesting genetic logic circuits or metabolic pathways.
Other upcoming activities include:
- Cell-free synthetic biology workshops and meetings run by early career researchers from the perspective of basic and applied research and education, particularly in an international development context.
- Student-led hands-on projects organised by the Cambridge University Synthetic Biology Society.
- A BBSRC Grand Challenges Research Fund supported meeting at the University of Pretoria, South Africa organised by OpenPlant and the Earlham Institute Foundry, which features cell-free systems as a major theme.
- Continuation of the Development i-Teams project to further prioritise and prototype applications of greatest relevance for improving the lives of people in the global South.
References
[1] Pardee, Keith, et al. "Paper-based synthetic gene networks." Cell 159.4 (2014): 940-954.
[2] Garamella, Jonathan, et al. "The all E. coli TX-TL Toolbox 2.0: a platform for cell-free synthetic biology." ACS synthetic biology 5.4 (2016): 344-355.

